

|
单细胞测序技术服务 靶向单细胞测序(lncRNA&mRNA) 单细胞测序 |
|
生物分子凝聚体研究 HyPro靶RNA临近标记技术 |
|
NGS测序技术服务 R-loop 测序(DRIPc-seq) |
|
NGS测序技术服务 环状DNA测序(eccDNA测序) |
|
Ribo-seq Ribo seq(ribosome profiling) |
核糖体-新生肽链复合物(RNC) RNC联合 circRNA芯片 RNC联合 lncRNA芯片 RNC-seq |
|
蛋白表达定量 Label free非标定量 TMT标记定量 PRM靶向定量 |
相关产品
Arraystar SYBR® Green Real-time qPCR Master Mix相关服务
miRNA测序相关资源
Arraystar nrStar™ Canonical Conserved miRNA PCR芯片(H)旨在通过qPCR技术检测372个经典的、进化过程保守且具有显著功能的mature miRNAs。此款PCR芯片能很好地消除miRNA前体、家族其他成员和isomiRs的干扰,同时还能够准确地对低量样本进行检测,为科研人员提供一种准确、简单和快捷的miRNA研究方案。
产品列表
| 产品名称 | 货号 | 规格 | 描述 |
|---|---|---|---|
| nrStar™ Human Canonical Conserved miRNA PCR Array | AS-NR-005 | 384 well / plate | 包含372个经典的、进化过程保守且具有显著功能的mature miRNAs |
| nrStar™ Human Canonical Conserved (Roche Light Cycler 480) | AS-NR-005-R | 384 well / plate |
优势
• 专注于经典、进化保守且具有显著功能的mature miRNAs
• 关注于具有更高表达水平的mature miRNAs而非star miRNAs
• 通过Two-tailed qPCR技术高效地消除前体、isomiRs及家族其他成员的干扰
• 详细地注释了miRNA启动子、初级转录本及宿主基因等信息
• 方便快捷的预扩增步骤帮助准确地检测低量样本
为保证准确地检测miRNA表达水平,消除前体、isomiRs及家族其他成员的影响,推荐使用rtStar™ First-Strand cDNA Synthesis Kit (3’and 5’adaptor) (Cat# AS-FS-003)进行cDNA合成。针对低量样本(100pg至200ng RNA),建议使用rtStar™ PreAMP cDNA Synthesis Kit (Cat# AS-FS-006)进行cDNA合成及预扩增步骤。
严谨的目标miRNA筛选收录流程

图1.严谨地筛选经典、进化保守的miRNAs用于PCR芯片检测。首先分析miRBase中所有pre-miRNA条目,挑选出经典pre-miRNA分子;其次将这些pre-miRNA进行系统发生学分析,找出进化过程中保守的miRNA;最后根据成熟miRNA的表达量,挑选出具有更高表达水平的mature miRNAs用于PCR芯片检测。
专注于经典的miRNAs
miRBase收录的miRNAs中,大约仅有1/3的人类miRNA基因、1/6的后生动物miRNA基因被认为是真实的miRNA基因(authentic miRNAs)[1]。经典miRNAs(Canonical miRNAs)拥有更为广谱的表达,同时也能在不同样本中具有差异性的表达(图2)[2]。反之,大部分的非经典miRNAs(Noncanonical miRNAs)具有很差的保守性以及更低的表达水平,这预示着它们不具备显著的基因表达调控能力[3]。故而,我们有理由在miRNA研究过程中关注于这些具有显著功能的miRNA类群——经典miRNAs。
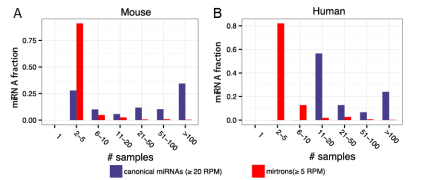
图2. 经典miRNAs及mirtrons在不同样本中的表达分布。
研究进化保守的miRNAs
与进化后期出现的miRNA基因相比,保守的miRNA(Conserved miRNAs)拥有更高的表达水平[3, 4]。在进化过程中新出现的miRNA基因往往对机体发挥中性亦或不利的调控功能,并且它们中的大部分在进化过程中慢慢地消失了[5]。然而保守的miRNAs的生物学功能则经历了进化筛选,是生物体正常运转所必需的。实际上保守的miRNAs能够调控比非保守miRNAs更多的靶基因,并且这些靶基因中很大一部分行使转录因子功能(图3)[6-8]。
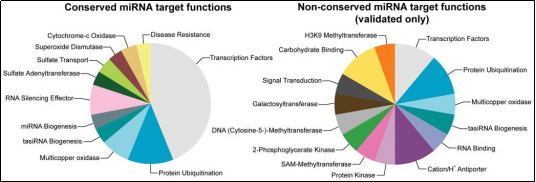
图3. 保守miRNAs(左)及非保守miRNAs(右)的靶基因功能统计分析。保守miRNAs主要调控转录因子的基因表达,而非保守miRNAs的靶基因功能则相对弥散。
聚焦于mature miRNAs而非star miRNAs
miRNA前体在Dicer酶的作用下产生双链RNA,一条指导链和一条随从链。当两条链的表达水平相差2倍以上时,更高的成为mature miRNAs,而另一条则为star miRNAs;如若两条链表达水平相当,则两条链均称为mature miRNAs[1]。再者,Star miRNAs发挥更为微弱的生物学功能。进化分析发现,star miRNAs拥有比mature miRNAs更高的碱基替代率,这种现象在seed区域显得更为明显(>46倍的差异)(图4)。
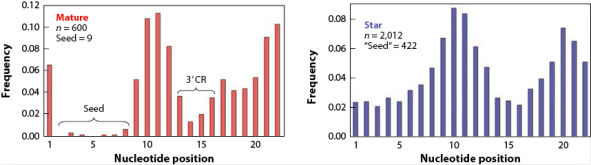
图4. Mature miRNAs和Star miRNAs的碱基替代率。n为所有碱基替代次数统计。
可区分前体、isomiRs及family members的two-tailed qPCR技术
由于miRNA有长度短小、isomiRs的存在、家族成员间高度同源以及特定样品中低表达丰度等特点,使得miRNA的表达检测显得更为困难。虽然qPCR作为miRNA表达检测中的“金标准”,然而传统的qPCR方法也不能够解决miRNA检测中的困难。这是由于qPCR方法依赖于引物3’末端的差异,然而对于5’端的差异则爱莫能助[9, 10]。而传统miRNA qPCR检测方法仅仅依赖一条上游引物进行区分,下游多为通用引物。而生命体内有众多RNAs与miRNAs在5’端存在单碱基差异,例如isomiRs以及同一家族其他miRNA成员。再者Seed序列位于5’区域,是miRNA识别靶基因的关键所在。Arraystar开发的two-tailed qPCR技术通过延伸miRNA两端,使得区分前体、isomiRs以及家族其他成员更为容易。再者,该技术能够方便快捷地进行低量样本(低至100pg RNA)的预扩增操作(图5)。
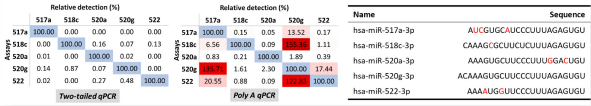
图5. 利用Two-tailed qPCR技术及Poly-A qPCR技术分别对人工合成的miR-515家族miRNAs进行交叉检测。
针对目标miRNAs进行全面注释
| Annotation | Implications |
|---|---|
| Tissue-specific | The miRNAs can have the tissue-specific functions in the corresponding tissue. |
| Cell-specific | Cell-type specific miRNAs tend to target proteins at crucial bottleneck positions in the corresponding regulatory networks. |
| Detection in Serum/plasma | miRNAs in serum/plasma having good potentials in biomarker applications. |
| Promoter name |
Information about the miRNA biogenesis. Also, many miRNA host genes are associated with the target gene activity either synergistically or antagonistically. |
| Promoter Annotation | |
| Primary miRNA | |
| Host Gene | |
| Intronic/intergenic | |
| Seed family | miRNAs in the same seed family are evolutionarily closer and have at least partially redundant functions. |
| Clustered miRNAs | miRNAs clustered in the genome generally have a similar expression level. They may regulate overlapping sets of target genes through convergent evolution. |
参考文献:
1. Fromm B, Billipp T, Peck LE, Johansen M, Tarver JE, King BL, Newcomb JM, Sempere LF, Flatmark K, Hovig E et al: A Uniform System for the Annotation of Vertebrate microRNA Genes and the Evolution of the Human microRNAome. Annual review of genetics 2015, 49:213-242.
2. Wen J, Ladewig E, Shenker S, Mohammed J, Lai EC: Analysis of Nearly One Thousand Mammalian Mirtrons Reveals Novel Features of Dicer Substrates. PLoS computational biology 2015, 11(9):e1004441.
3. Bartel DP: Metazoan MicroRNAs. Cell 2018, 173(1):20-51.
4. Nozawa M, Miura S, Nei M: Origins and evolution of microRNA genes in Drosophila species. Genome biology and evolution 2010, 2:180-189.
5. Meunier J, Lemoine F, Soumillon M, Liechti A, Weier M, Guschanski K, Hu H, Khaitovich P, Kaessmann H: Birth and expression evolution of mammalian microRNA genes. Genome research 2013, 23(1):34-45.
6. Agarwal V, Bell GW, Nam JW, Bartel DP: Predicting effective microRNA target sites in mammalian mRNAs. eLife 2015, 4.
7. Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB: Prediction of mammalian microRNA targets. Cell 2003, 115(7):787-798.
8. Fahlgren N, Howell MD, Kasschau KD, Chapman EJ, Sullivan CM, Cumbie JS, Givan SA, Law TF, Grant SR, Dangl JL et al: High-throughput sequencing of Arabidopsis microRNAs: evidence for frequent birth and death of MIRNA genes. PloS one 2007, 2(2):e219.
9. Git A, Dvinge H, Salmon-Divon M, Osborne M, Kutter C, Hadfield J, Bertone P, Caldas C: Systematic comparison of microarray profiling, real-time PCR, and next-generation sequencing technologies for measuring differential microRNA expression. Rna 2010, 16(5):991-1006.
10. Lefever S, Pattyn F, Hellemans J, Vandesompele J: Single-nucleotide polymorphisms and other mismatches reduce performance of quantitative PCR assays. Clinical chemistry 2013, 59(10):1470-1480.
适用的qPCR仪:
ABI ViiA™ 7
ABI 7500 & ABI 7500 FAST
ABI 7900HT
ABI QuantStudio™ 6 Flex Real-Time PCR system
ABI QuantStudio™ 7 Flex Real-Time PCR system
ABI QuantStudio™ 12K Flex Real-Time PCR System
Bio-Rad CFX384
Bio-Rad iCycler & iQ Real-Time PCR Systems
Eppendorf Realplex
QIAGEN Rotor Gene Q 100
Roche LightCycler 480
Stratagene Mx3000
Roche LightCycler 480
|
Human Canonical Conserved miRNA列表 (372 miRNAs) |
|
非常高度进化保守miRs (6): miR-31-5p, miR-96-5p, miR-182-5p, miR-184, miR-210-3p, miR-375-3p 高度进化保守miRs (162): let-7a-5p, let-7b-5p, let-7c-5p, let-7d-5p, let-7e-5p, let-7f-5p, let-7g-5p, let-7i-5p, miR-1-3p, miR-7-5p, miR-9-5p, miR-10a-5p, miR-10b-5p, miR-15a-5p, miR-15b-5p, miR-16-5p, miR-17-5p, miR-18a-5p, miR-18b-5p, miR-19a-3p, miR-19b-3p, miR-20a-5p, miR-20b-5p, miR-21-5p, miR-22-3p, miR-23a-3p, miR-23b-3p, miR-24-3p, miR-25-3p, miR-26a-5p, miR-26b-5p, miR-27a-3p, miR-27b-3p, miR-29a-3p, miR-29b-3p, miR-29c-3p, miR-30a-5p, miR-30b-5p, miR-30c-5p, miR-30d-5p, miR-30e-5p, miR-32-5p, miR-33a-5p, miR-33b-5p, miR-34a-5p, miR-34b-5p, miR-34c-5p, miR-92a-3p, miR-92b-3p, miR-93-5p, miR-98-5p, miR-99a-5p, miR-99b-5p, miR-100-5p, miR-101-3p, miR-103a-3p, miR-106a-5p, miR-106b-3p, miR-106b-5p, miR-107, miR-122-5p, miR-125a-5p, miR-125b-5p, miR-126-3p, miR-126-5p, miR-128-3p, miR-129-5p, miR-130a-3p, miR-130b-3p, miR-132-3p, miR-133a-3p, miR-133b, miR-135a-5p, miR-135b-5p, miR-137-3p, miR-138-5p, miR-139-5p, miR-140-3p, miR-141-3p, miR-142-3p, miR-142-5p, miR-143-3p, miR-144-5p, miR-145-5p, miR-146a-5p, miR-146b-5p, miR-147b-3p, miR-148a-3p, miR-148b-3p, miR-150-5p, miR-152-3p, miR-153-3p, miR-155-5p, miR-181a-5p, miR-181b-5p, miR-181c-5p, miR-181d-5p, miR-183-5p, miR-187-3p, miR-190a-5p, miR-191-5p, miR-192-5p, miR-193a-5p, miR-193b-3p, miR-193b-5p, miR-194-5p, miR-195-5p, miR-196a-5p, miR-196b-5p, miR-199a/b-3p, miR-200a-3p, miR-200b-3p, miR-200c-3p, miR-202-3p, miR-202-5p, miR-203a-3p, miR-204-5p, miR-205-5p, miR-206, miR-208a-3p, miR-208b-3p, miR-211-5p, miR-212-5p, miR-214-3p, miR-215-5p, miR-216b-5p, miR-217-5p, miR-218-5p, miR-219a-2-3p, miR-219a-5p, miR-221-3p, miR-222-3p, miR-223-3p, miR-301a-3p, miR-301b-3p, miR-302a-3p, miR-302b-3p, miR-302c-3p, miR-302d-3p, miR-338-3p, miR-363-3p, miR-365a-3p, miR-383-5p, miR-424-5p, miR-425-5p, miR-429, miR-449a, miR-449b-5p, miR-449c-5p, miR-451a, miR-454-3p, miR-455-3p, miR-455-5p, miR-489-3p, miR-490-3p, miR-497-5p, miR-499a-5p, miR-503-5p, miR-551a, miR-551b-3p, miR-802, miR-875-5p 进化保守miRs (204): miR-28-3p, miR-105-5p, miR-127-3p, miR-134-5p, miR-136-3p, miR-149-5p, miR-151a-3p, miR-151a-5p, miR-154-3p, miR-154-5p, miR-185-5p, miR-186-5p, miR-188-5p, miR-197-3p, miR-224-5p, miR-296-5p, miR-299-3p, miR-299-5p, miR-323a-3p, miR-323b-3p, miR-324-5p, miR-326, miR-329-3p, miR-330-3p, miR-331-3p, miR-335-5p, miR-337-3p, miR-339-3p, miR-339-5p, miR-340-5p, miR-342-3p, miR-345-5p, miR-346, miR-361-3p, miR-361-5p, miR-362-5p, miR-369-3p, miR-369-5p, miR-370-3p, miR-371a-5p, miR-372-3p, miR-373-3p, miR-374a-3p, miR-374a-5p, miR-374b-5p, miR-376a-3p, miR-376b-3p, miR-376c-3p, miR-377-3p, miR-377-5p, miR-378a-3p, miR-379-5p, miR-380-3p, miR-380-5p, miR-381-3p, miR-382-3p, miR-382-5p, miR-409-3p, miR-410-3p, miR-411-5p, miR-412-5p, miR-421, miR-423-3p, miR-423-5p, miR-431-5p, miR-432-5p, miR-433-3p, miR-448, miR-450a-5p, miR-450b-5p, miR-452-5p, miR-483-5p, miR-485-5p, miR-486-5p, miR-487a-5p, miR-487b-3p, miR-488-3p, miR-488-5p, miR-491-5p, miR-493-5p, miR-494-3p, miR-495-3p, miR-496, miR-498-5p, miR-500a-3p, miR-500b-5p, miR-501-3p, miR-502-3p, miR-504-5p, miR-506-3p, miR-506-5p, miR-507, miR-508-3p, miR-510-5p, miR-511-3p, miR-511-5p, miR-513b-5p, miR-514a-3p, miR-514b-5p, miR-516b-5p, miR-518b, miR-518c-3p, miR-518d-5p, miR-518f-3p, miR-519c-3p, miR-520a-3p, miR-520d-3p, miR-522-3p, miR-523-3p, miR-524-3p, miR-524-5p, miR-525-3p, miR-525-5p, miR-532-5p, miR-539-3p, miR-541-5p, miR-542-3p, miR-543, miR-545-5p, miR-574-3p, miR-576-5p, miR-577, miR-579-5p, miR-582-3p, miR-584-5p, miR-589-5p, miR-590-3p, miR-597-3p, miR-598-3p, miR-605-3p, miR-624-5p, miR-628-5p, miR-651-5p, miR-652-3p, miR-654-3p, miR-656-3p, miR-660-5p, miR-671-3p, miR-671-5p, miR-675-3p, miR-676-3p, miR-708-5p, miR-744-5p, miR-758-3p, miR-760, miR-769-5p, miR-770-5p, miR-873-3p, miR-873-5p, miR-874-3p, miR-876-3p, miR-885-5p, miR-887-3p, miR-887-5p, miR-888-5p, miR-889-3p, miR-890, miR-892a, miR-892b, miR-892c-3p, miR-934, miR-935, miR-944, miR-1180-3p, miR-1185-1-3p, miR-1185-2-3p, miR-1185-5p, miR-1197, miR-1247-5p, miR-1249-3p, miR-1251-5p, miR-1264, miR-1271-5p, miR-1277-5p, miR-1283, miR-1298-5p, miR-1301-3p, miR-1307-3p, miR-1307-5p, miR-1323, miR-1343-3p, miR-1911-5p, miR-1912-3p, miR-2114-3p, miR-2114-5p, miR-2355-3p, miR-2681-3p, miR-2681-5p, miR-3059-5p, miR-3085-3p, miR-3140-3p, miR-3145-3p, miR-3146, miR-3173-5p, miR-3200-3p, miR-3613-5p, miR-3617-5p, miR-3934-5p, miR-4524a-3p, miR-4677-3p, miR-4766-3p, miR-4766-5p, miR-6529-5p, miR-9851-3p |

